How to select the right profile for your resource
In this how-to, we will guide you through the necessary steps for you to select a Bioschemas profile that will be later used to add mark up to your own resources
1. Your first encounter with Bioschemas profiles
You can find the availabe Bioschemas profiles at http://bioschemas.org/specifications. There, you will be presented with a list of all the current and stable profiles, as illustrated on Figure 1. You can hover on the profile name to see a quick description. Should you need a more detailed information, just click on the profile name.
As seen on Figure 1, each profile will show you details such as current version, release date, use cases, crosswalk, tasks and issues, usage examples and current live deploys.
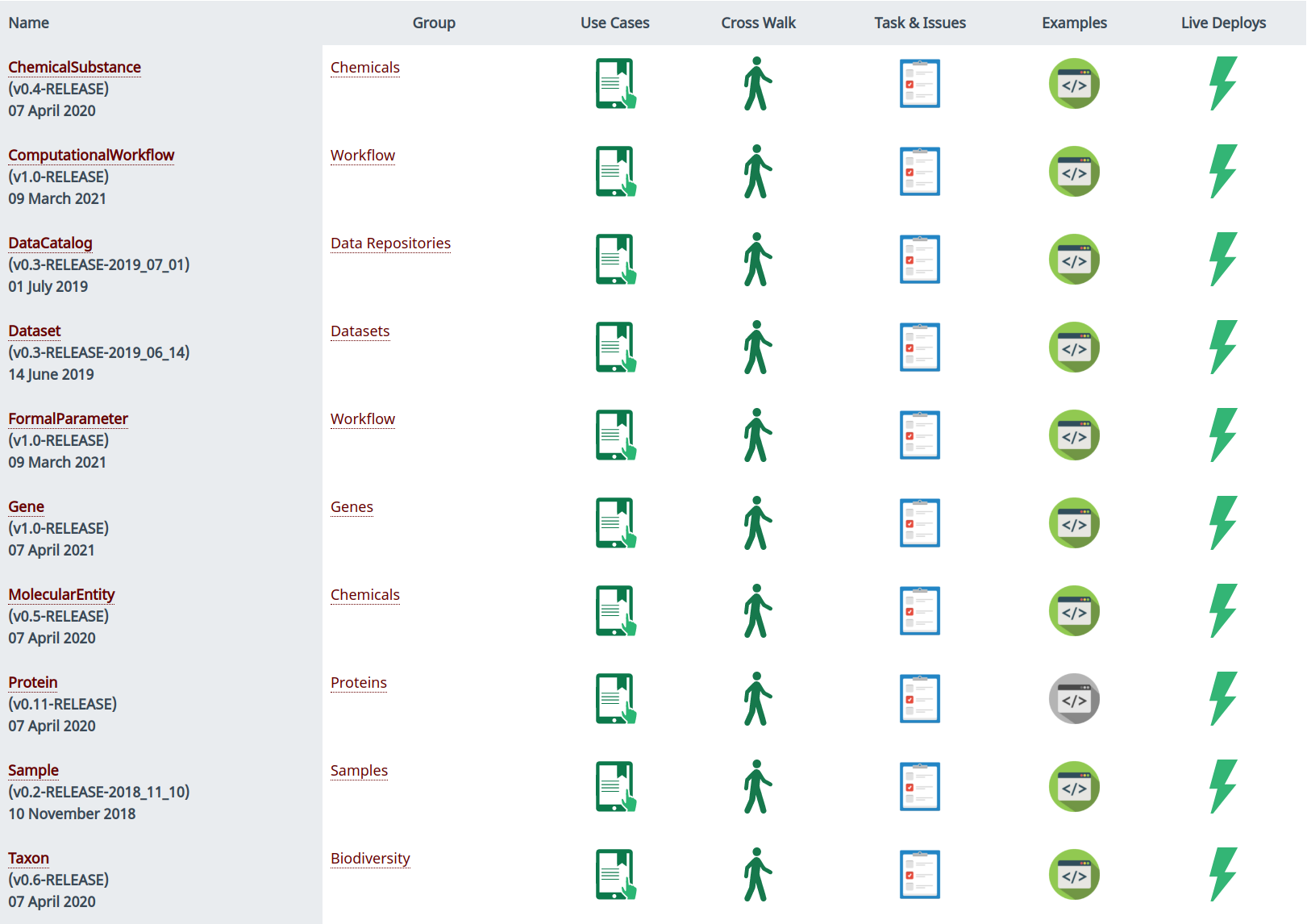

- Use cases: Used as a basis for the profile
- Crosswalk: Documentation on the brainstorming and process followed by a group in order to come up with a profile specification
- Tasks & issues: Link to a GitHub space where you can report issues with a profile, see the assignees, and participate of the discussion
- Example: Usage examples for the profile
- Live deploys: Link to live deploys for the profile
2. Pick up a profile
Which profile is the right one for you will depend on your resource. Go through the list above, and try to figure out which one most closely matches your use case. If you cannot find any relevant profile, then check out the “Drafts” tab where some new profiles or new profile releases are being discussed.
If you still cannot find any profile suited for your needs, do not hesitate to engage with the community by submitting an issue on Github.
In the next section, we will present some hints on those profiles that have been more broadly used so far, i.e., mainly customizing generic types rather corresponding to specific Life Science entities.
3. A guided tour to some selected Bioschemas profiles
3.1. DataCatalog
Also known as data repository, a data catalog commonly aggregates more than one dataset. If your resource supports only one dataset, you still could decide to markup your resource, in this case, as DataCatalog and also Dataset (this would make it easier if you are thinking of adding more datasets. However, whenever more than one dataset is provided, it totally makes sense to markup your resource as a DataCatalog.
3.2. Dataset
If your resource provides data and you can easily identify a common entity type for all the data contained in it, you should probably go for a Dataset profile. Let’s clarify what we mean by “common type”. Let’s suppose you have chemical compounds including drugs, proteins and cells. If you see them all as the same thing, chemical compound, you have one Dataset, and you have found the right profile for you. However, if you actually distinguish drugs from proteins from cells and so, and (maybe even) tailor the information provided for each case, you have a data catalog and multiple datasets, you should use both, one DataCatalog and multiple Datasets.
3.3. MolecularEntity
When your webpages describe molecules then you can identify them with the MolecularEntity profile. This profile allows you to give details on every molecule separately, unlike with Dataset which describes normally many molecules. The MolecularEntity allows you to specify molecule-specific information like an InChI or InChIKey, IUPAC name, molecular formula, and even the SMILES line notation for the molecule.
3.4. ChemicalSubstance
When the chemical is not a well-defined molecule but, for example, a nanomaterial, then you can annoated them with the ChemicalSubstance profile. This profile allows you to give details on the identifier of the substance and a chemical composition. The profile inherits properties from BioChemEntity, like chemical and biological roles.
3.5. ComputationalTool
If your webpage describes software aimed at producing or analyzing scientific data, then you can annotate it with the ComputationalTool profile. This profile guides you on providing metadata from the SoftwareApplication type, such as the tool name and a short description. In addition, thanks to the EDAM ontology, you can provide semantic annotations to document what the tool does, what kind of data is taken as input, or produced as ouput.
3.6. ComputationalWorkflow
When you are publishing on the web a complex data analysis, made of several processing steps, you can annotate it with the ComputationalWorkflow profile. This profile provides guidelines to choose metadata inherited from the SoftwareSourceCode type. For instance the community agreed to document, among other minimal metadata, workflow input and output parameters as well as the workflow license.
3.7. Course, CourseInstance and TrainingMaterial
These three profiles work well together but also on their own. Whether you use them all together or not, depends on your own use case. Here we present the different scenarios and things to consider.
A Course is used to describe the broad, common aspects of a recurring training event (e.g., a course “Introduction to Bioinformatics” part of the regular delivery of a Bionformatics master in a university), whereas a CourseInstance is about the specific times and location of when a particular version of a Course is held (e.g., the specific edition of that “Introduction to Bioinformatics” course on the first academic semester of 2022). Course and CourseInstance have been design to be used in tandem. These two views on a training event, i.e., Course and CourseInstance, are complemented with TrainingMaterial (aka LearningReource in Schema.org). Below we present a series of scenarios that should help you decide the right training profile for your own case.
Scenario 1 - Course alone A new master in “Biomedical Data Science” is starting in the following semester and the delivery includes some optional courses that will be scheduled depending on the demand, e.g., “Description logics for Biomedical Ontologies”. At this point, it has not beed decided when exactly the first edition of this course will run. In this case, you have a Course but you do not have (yet) a CourseInstance or TrainingMaterial.
Scenario 2 - One Course and one CourseInstance An “Introduction to Bioschemas” course will be offered as part of the tutorials of the “Bionformatics Amazing Conference 2022”, this course (aka tutorial in this scenario) is scheduled for one day, on the 13th of May of 2022. At this point you do not know if the same course will ever be run again. In this case, you have one Course “Introduction to Bioschemas” with one CourseInstance offered at the mentioned conference.
If it is one CourseInstance only, cannot I not simply use CourseInstance and forget about Course? No, a CourseInstance should always be used in tandem with a Course. Course includes elements such as description and name which are not included in CourseInstance. You would use Course for those “generic” aspects of a course (e.g., its name and description) while CourseInstance for elements spefic to a particular edition (e.g., location).
Scenario 3 - A course, at least one CourseInstance and at least one TrainingMaterial In addition to what you had in Scenario 2, you also have the training material corresponding to the first half where you explain “What is Bioschemas about?” Your material has a name “What is Bioschemas about?”, a description and some keywords, in addition, you have also defined some pre-requisites aka competency required for learners/trainees (e.g., basic knowledge on Schema.org), and the target audience (e.g., bioinformaticians). In this case, you have the Course and CourseInstance from Scenario 2 but also a *TrainingMaterial”.
As an example (or learning activity), I am going to use a protein dataset to show how to use the Bioschemas profile Dataset, does it mean that that protein dataset becomes a “TrainingMaterial”? Not really. The protein dataset remains a Dataset that happens to be momentarily used for a training activity, i.e., as part of a TrainingMaterial. We can use TraininMaterial mentions “Dataset” to make the connection explicit, and give credit to the dataset that we are reusing and make it easier for others to reach that dataset.
Scenario 4 - A stand-alone TrainingMaterial As part of the Bioschemas website, we offer to the community a tutorial on Adding Schema.org to a GitHub Pages site. We do not know when this tutorial will be used, by whom, or how long it will take for someone to go through it. We have, however, designed that page as a tutorial, i.e., a TrainingMaterial, that anyone can use on their own. In this case, we have a TrainingMaterial but no Course or CourseInstance.
3.8. Taxon, TaxonName
These two profiles can be used together or separately depending on your use case.
Taxon
A taxon is a group of organisms with common characteristics, identified by taxonomists to form a certain unit. If your resources deal with groups of biological organisms like animals, plants, fungi or bacteria, then you may want to use the Taxon profile.
The least you need to know to describe a Taxon is its full scientific name (i.e. including authorship and date), also called accepted name in botanics or valid name in zoology, and its taxonomimc rank such as species, genus, family etc. You may also provide synonyms of the accepted/valid scientific name that keep track of the taxonomic changes and thus reflect the way biologists think of a given taxon, that is, its circumscription.
For instance, the Beluga species has a valid scientific name which is Delphinapterus leucas (Pallas, 1776), a synonym name is Balaena albicans Muller, 1776, and its rank is species.
It is also a grood pratice to link a Taxon to its counterparts in third-party taxonomic registers like the NBCI taxonomy, or data agregators like GBIF or Catalog of Life so that people would know exactly what taxon you refer to.
TaxonName
Furthermore, some databases databases (e.g. Zoobank, IPNI and Mycobank) describe specifically scientific names but do not keep track of how names are assigned to taxa, such that they would be poorly supported by the use of the Taxon profile. In such a case, the TaxonName profile is more relevant to markup the resources of these taxonomic names registries.
Furthermore, the TaxonName profile may be used jointly with the Taxon profile when you want to describe a taxon while giving details about the scientific names.
How do I decide which one to use?
If you are unsure whether your resources deal with taxa or taxon names, just follow this simple rule:
- When your resources are primarily about biology in the broad sense, organisms, biological features, life traits, phenotypes etc., then most likely you should use the Taxon profile.
- When your resources relate to the publication and typification of scientific names, specifically when this pertains to the application of a Code of nomenclature, then you should use the TaxonName profile.
Keywords: schemaorg, markup, structured data, bioschemas profile
Topics:
Audience:
- (Markup provider, Markup consumer) People interested in selecting a Bioschemas profile to markup their own data
Authors:
Contributors:
License: CC-BY 4.0
Version: 2.1
Last Modified: 23 August 2022

