Use Cases for Scholarly Publication
The following use cases were developed by the Scholarly Publications group.
Biotea 2 Bioschemas - Introduction
Biotea defines a model for metadata, references, content and named entities recognized in the text. Bioschemas profiles for scholarly publications focus on metadata only.
How to cite Biotea:
- Model: https://jbiomedsem.biomedcentral.com/articles/10.1186/2041-1480-4-S1-S5
- Use cases: https://peerj.com/articles/4201/
Biotea model for metadata at a glance
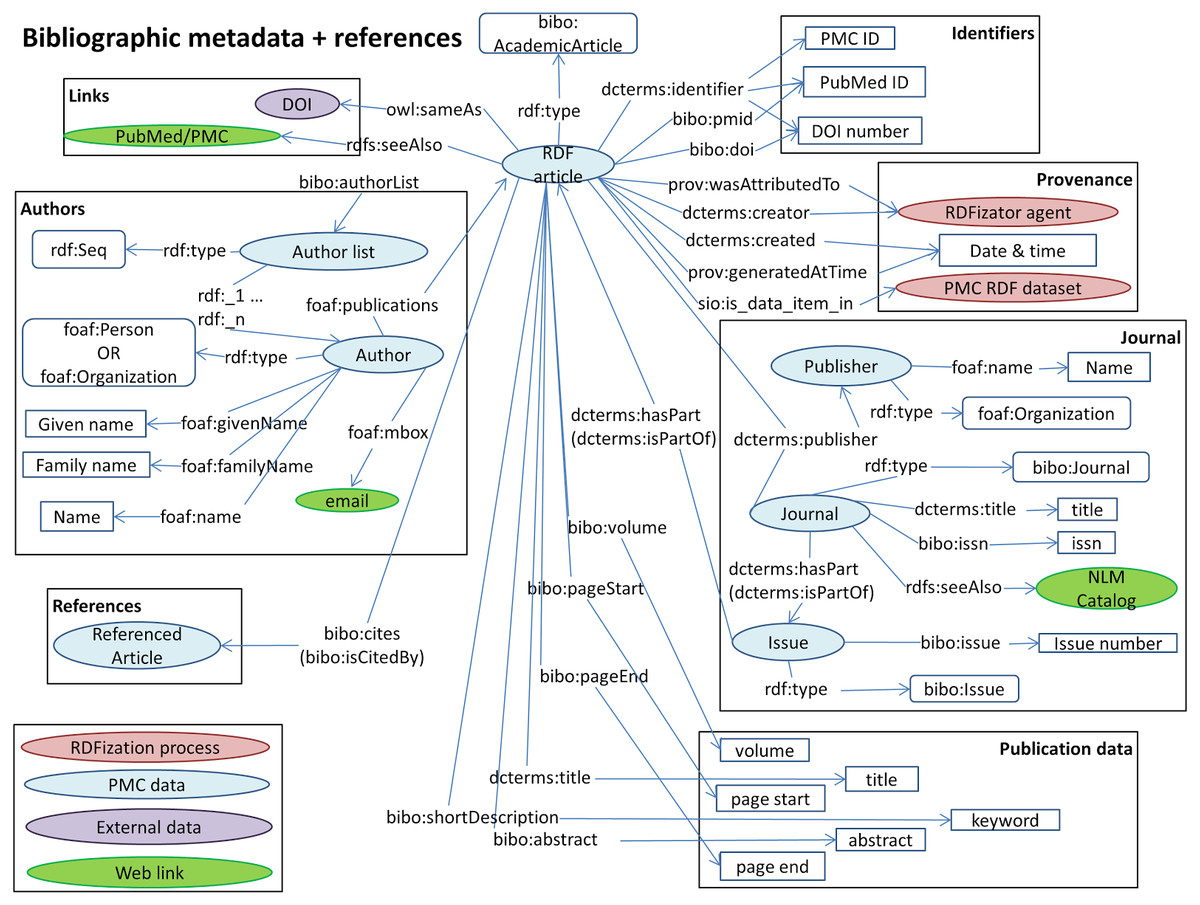
Use cases
The following use cases were derived from Biotea but they apply in general to search and retrieval of literature metadata and associated entities. A user here is anyone looking for literature related metadata and associated entities, e.g., a researcher trying to find articles about a particular protein or a researcher looking for literature metadata in a standard format so they can create a literature metadata parser or aggregator.
Findability
As a user, I want to easily find scholarly publications available on the web, regardless of where they are all hosted. As a user, whenever possible, I also want to find biological entities recognized in the text and use these to discover scholarly publications about the entity.
Bioschemas markup can enable an extended version of Google Scholar, where even institution hosted documents can be found, as far as they are marked up following the Scholarly Publication profiles.
Accessibility
As a user, I want to get literature metadata in a standard format, regardless of whether all articles are hosted on the same website or provide a specialized API.
As publications are marked up with Schema.org/Bioschemas, their key elements can be easily accessible by people and machines. Currently, the Journal Article Tag Suite is the default format supported by the main publishers in the Life Sciences domain. The JATS XML format is a comprehensive one including metadata, abstract, content and references. The format though lacks support for Linked Data. JSON is also supported via web services by some repositories such as PubMed – see e-utils tools, and Europe PMC, see RESTful services. Biotea adds a semantic layer on top of JATS as well as an easy way to access content, via annotations, with no need to expose it all.
Summarization
As a user, I want to see a summary similar to those for movies when I search for articles. This would become possible thanks to Schema.org/Bioschemas markup, providing a quick view of the publication.
Imagine you go to Google Scholar or Wikidata (or any other place aggregating scholarly publications metadata (it could be your institute or your own aggregating tool), thanks to Bioschemas markup, such websites could also include a summary of, for instance, citing publications on the last 3 months!
Interoperability
As a user, I would like to know what publications involve proteins; even more, I would like to know what proteins are mentioned in the text.
By adding some “about” data to literature markup, e.g., proteins, it could be possible to filter publications by the BioChemEntity they mention.

